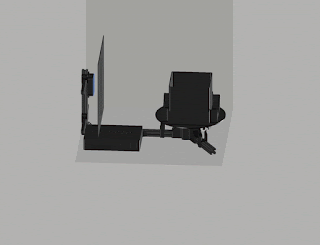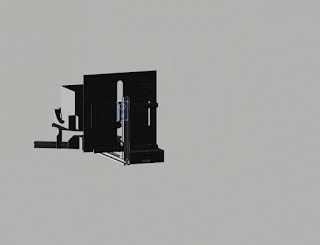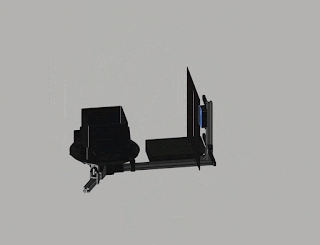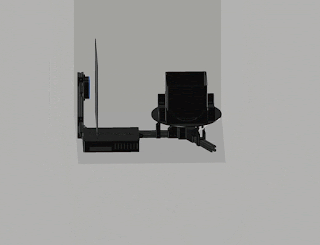
Robotic arm with a small foot-print
Master's degree/engineering course project
 |
Fig. 1. Six days of time-lapse tracking of the root growth. Arabidopsis thaliana seedlings
grown on a Petri dish in a plant growth chamber. |
The research in our
group is focused on molecular mechanisms underpinning development of plants. It is similar to engineering when
there is nobody around to explain you what all the parts are for. So, on daily
basis, we break plant genes to see what they were needed for. To track plant growth
properly we need a good camera system that will image plants for days or even
weeks (exactly like Fig 1, but less
blurry). We grow our plants in small Petri dishes within a growth cabinet and
thus need the imaging platform to have small foot print.
Preliminary results:
We put
together the minimal version of the imaging stage (see Fig. 2).
Despite the questionable quality of the design, it was such a significant
improvement of our experiments that we decided to invest into constructing a
proper system. Our system is built around a Raspberry Pi computer, connected to
a camera capable of producing images both under daylight conditions and at
night using near-infrared illumination.
Fig. 3. Update (August 2018). No more double-sided tape on cardboard! We are switching to 3D printing.
Project goals and requirements:
1. Make the imaging stage less prone to falling apart.
Additionally, the stage should be constructed in such a way that it provides
more consistent images with regards to lighting.
2. Develop a
small footprint robot that can move plates from the growth position to the
imaging stage and back. This will enable parallel imaging of multiple plates,
while not compromising the amount of light available to the plants.
3. The robot
needs to be able to communicate with the Raspberry Pi, either via ethernet,
WiFi, bluetooth or serial/GPIO (or you may have other suggestions).
 |
Fig 3. The team.
|
3. We will introduce you to engineering on the DNA level.